Q: What is inbreeding?
A: Each animal receives two gametes, one from the sire and the other from the dam. Therefore, an animal has a maternal and a paternal copy of each gene. If the two copies of the gene are the same, the animal is homozygous
for that specific location and if the homozygosity can be traced to a common ancestor(s) the animal is inbred for that location. This is known as identity by descent or IBD. The average of IBD across the whole genome is quantified in a value called
the inbreeding coefficient. The inbreeding coefficient represents the overall inbreeding of the animal and is the likelihood that the maternal and paternal copy of each allele are identical by descent. Let’s define some terms to have a better
understanding of inbreeding. 
Allele: Each gene may have different variants with different functions. These variants are called alleles.
Homozygosity: A locus is homozygous when the two alleles of a gene are identical. A homozygote locus can be identity by state (IBS) or identity by descent (IBD).
Identity by state: When the two alleles are identical by chance.
Identity by descent: When the two alleles are identical because they are copies of the same ancestral allele.
In inbreeding context, the IBD is what we measure which is the result of mating between related animals.
An individual inherits two alleles for each gene; one from the sire and one from the dam. Alleles are located at the same position within homologous chromosomes.
Homozygous: If both alleles are identical, the individual is homozygous for this gene.
Heterozygous: If both alleles are different, the individual is heterozygous for this gene.
Q: How is inbreeding measured?
A: Inbreeding can be calculated from the pedigree or the genotypes of animals.
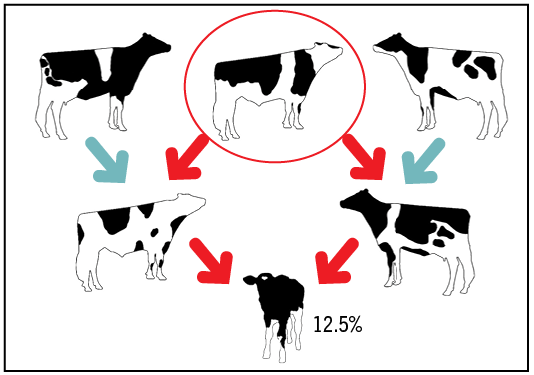 Pedigree-based inbreeding: In this method, probability of IBD is calculated from the known ancestry. The inbreeding is a function of how many common ancestors are in the pedigree of the animals and how many generations
there are between the common ancestors.
Pedigree-based inbreeding: In this method, probability of IBD is calculated from the known ancestry. The inbreeding is a function of how many common ancestors are in the pedigree of the animals and how many generations
there are between the common ancestors.
Pros: The method is simple; no genotype is needed.
Cons: Pedigree completeness and errors affect the accuracy. The variation in gene segregation during reproduction is not fully considered, for example full siblings will have the same inbreeding with this method.
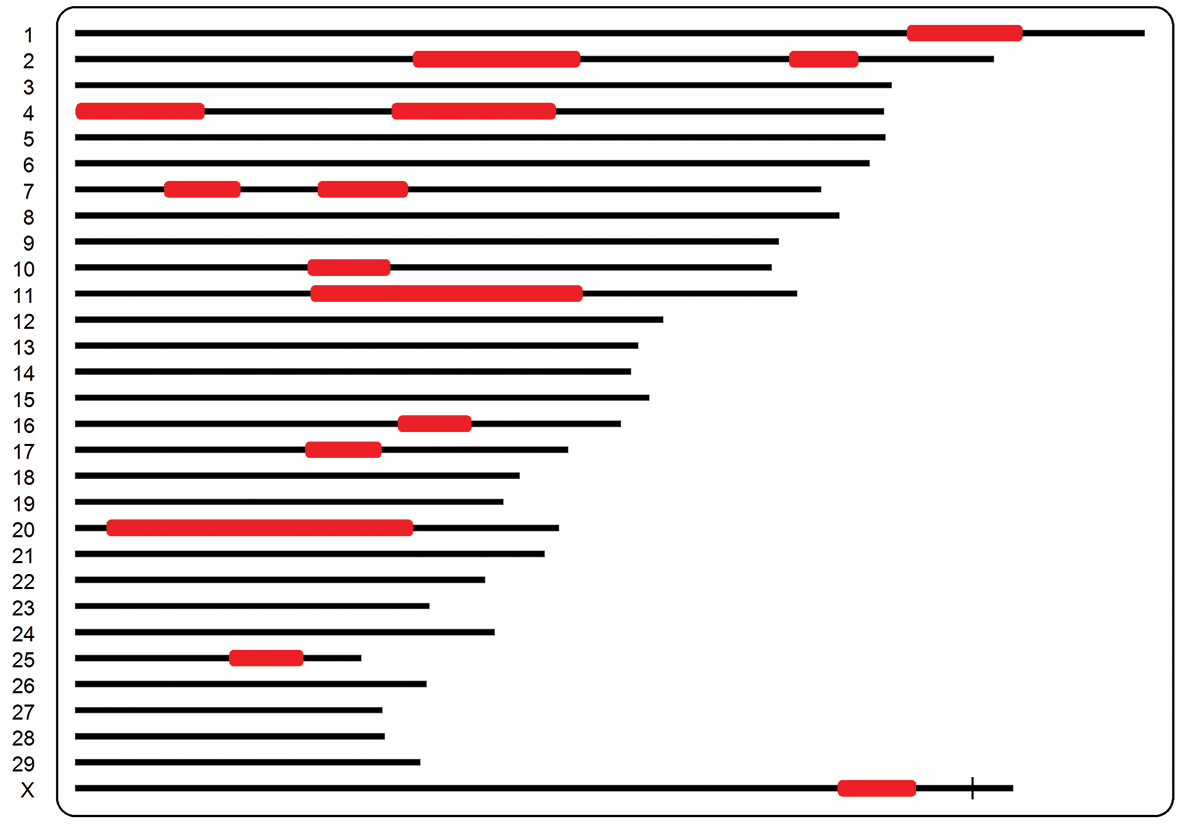 Genomic-based inbreeding: In this method, observed genotypes (covering the whole genome uniformly) are used to calculate IBD. There are different approaches to calculate inbreeding from genotypes. A common method
uses runs of homozygosity (ROH) which are long segments of homozygous loci in the genome. ROH is more accurate than pedigree-based inbreeding because it accounts for variations in gene segregation.
Genomic-based inbreeding: In this method, observed genotypes (covering the whole genome uniformly) are used to calculate IBD. There are different approaches to calculate inbreeding from genotypes. A common method
uses runs of homozygosity (ROH) which are long segments of homozygous loci in the genome. ROH is more accurate than pedigree-based inbreeding because it accounts for variations in gene segregation.
Pros: Pedigree information is not required and therefore pedigree errors have no impact on ROH. Gene segregation differences are considered, for example full siblings may have different inbreeding with this method. Inbreeding coefficients
can be calculated for each chromosomal region which allows separating out high- and low-risk inbreeding.
Cons: This method is more complex, and genotypes are needed. Genotyping errors may influence the calculation but, in general, the genotyping error rate is very low.
Q: What is the impact of inbreeding?
A: An elevated level of inbreeding in the individual/population can lead to reduced fitness and consequently lower performance known as inbreeding depression.
Q: What is the acceptable level of inbreeding?
A: There is no magical threshold for inbreeding as the improved environment can mask the negative impact of inbreeding to some degree. That means an inbred animal with no genetic disorder may not show significant
inbreeding depression in a herd with good environmental management.
Q: How can we control inbreeding?
A: As a rule of thumb, use a diverse group of sires in the herd. Never let a few sires dominate the herd’s genetics. Use mating software to optimize mating based on your production goals while minimizing
the genomic/pedigree relationships between the sires and dams. Please note that relationships between service sires should also be monitored.
Take home message: ROH-based inbreeding is more accurate than pedigree-based inbreeding. With ROH, higher risk inbreeding (originating from recent common ancestors) can be identified. Sire diversification and mating software are effective ways to control the rate of inbreeding.